I have a 10GB root file with 30 columns and 60 million rows.
I got an error when I tried to import all data at once in pandas dataframe. I managed it by reading it in chunks, performing some pre selection cut, and managing data types.
After all that, I am finished with 2GB dataframe.
But still, when I tried to fit data to let’s say logistic regression or Random Forest, my kernel keeps dying/restarting.
Any solution?
Hi Toni,
It might still be that you are running out of memory, is there some copy going on?
There is one thing you can try: one of the SWAN machines (https://swan006.cern.ch) allows you to pick a maximum of 16 GB of memory when starting your session. Can you try that and see if you still get the kernel dying error?
On the other hand, I recommend that you have a look at ROOT's RDataFrame and its feature to convert to pandas:
https://root.cern/doc/master/df026__AsNumpyArrays_8py.html
You can process the dataset in your ROOT file with RDataFrame (filter it, define new columns, etc.) and then convert the columns you are interested in into Numpy arrays, which can be fed to pandas to create a dataframe. The good thing about this approach is that RDataFrame will not load all the dataset in memory to process it, so you will likely not experience the memory error issue.
Hi Enric, thank you for your reply.
Apropos your first proposition, i am already using SWAN006 with 16GB or RAM.
After I load my data, as I said, I am dealing with 2GB dataframe, which is not unusual;in past I have used even larger, so there is my concern.
I will definitely take a look at RDataFrame, and be back with response.
BR
Update.
I run same notebook locally on my laptop, and it went fine, without any kernel restart.
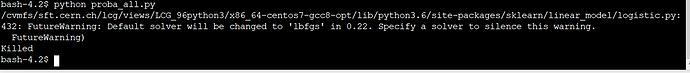
So it must be something about SWAN. When I tried to run pure python file in SWAN terminal, i got “killed” error.
Hi,
If you run the code from the Python prompt at the terminal, what is the error message that you see?
I see, can you narrow it down to a specific line in your code that causes the process to be killed?
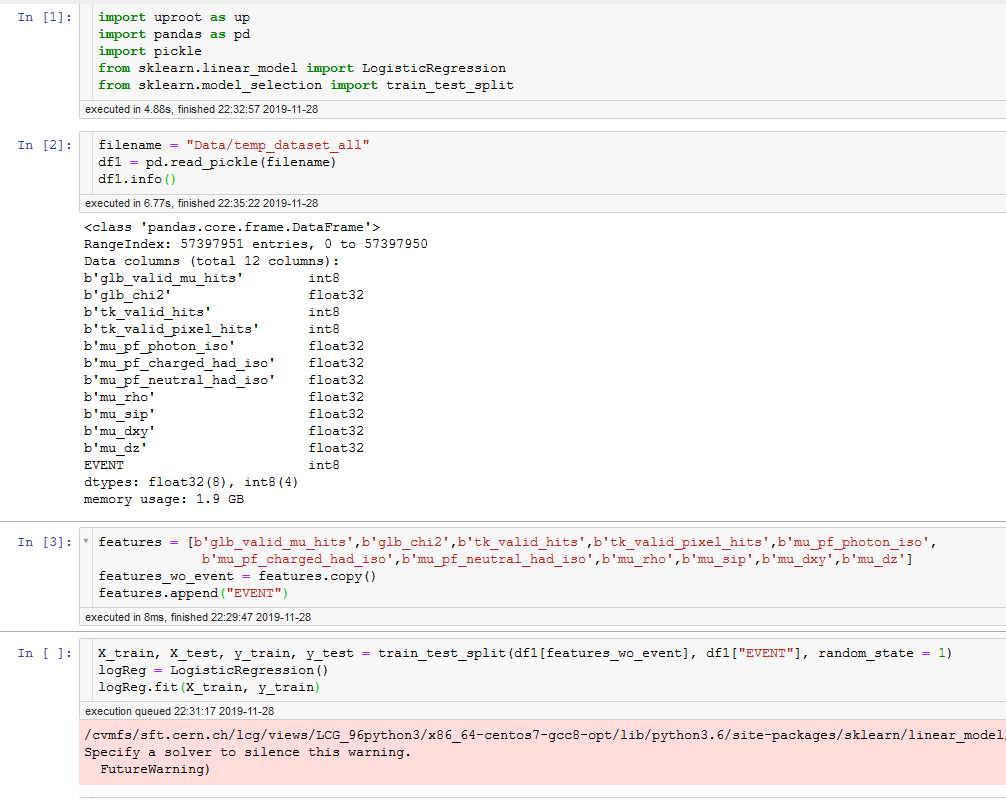
X = df1[features_wo_event].values
y = df1[“EVENT”].values
y = y.ravel()
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state = 1)
logReg = LogisticRegression()
> logReg.fit(X_train, y_train)
Hi Toni,
well, if you try to read in the whole 8GB file you’ll need at least 8 GB of memory (there will be some overhead, so it likely needs more), which any recent laptop provides. A shared system like SWAN needs to be more restrictive concerning the memory used per user, so you will only be allowed to use less than that, hence you won’t be able to do that on a “normal” SWAN instance – and maybe also not on lxplus (depending on the setup there) – and you get “killed” because your process runs out of memory. No magic here.
The only way out of that is to find a way to not read in the full file, like it’s done in RDataFrame.
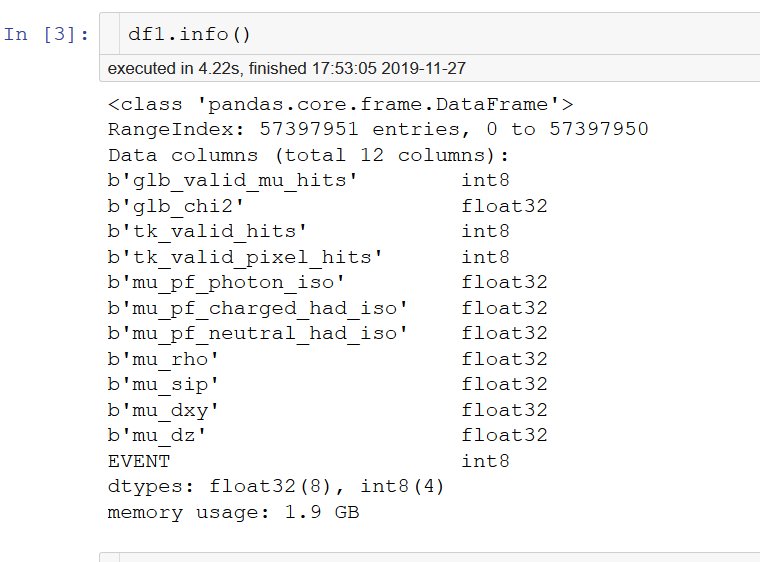
I understand. But, I made preselection cut during reading data in chunks, and as you can see on the picture, my final pandas DataFrame that I work with is 1.9 GB.
Edit: I also tried running same script on my older laptop(8GB of ram) and kernel also dies.
OK, so this DataFrame is about 2GB – did you make sure that the other objects you had during the creation are really removed from memory in the session ? The 12 columns with your intermediate results will add another ~2GB, and the last of object of your “chunked” read will also add to the total.
Since it crashes on your laptop with 8GB, it seems that some other items/objects still use some memory and the total is just too much … 
Can train_test_split be making a copy?
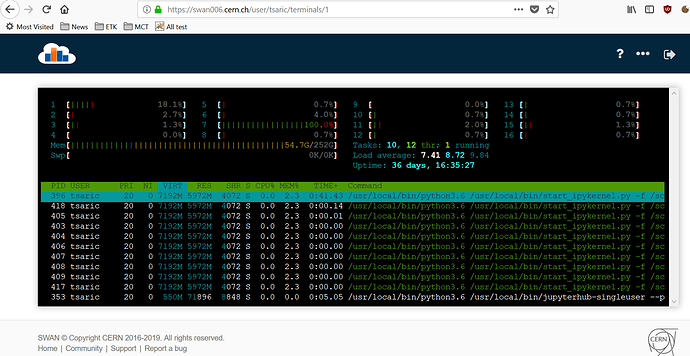
In SWAN, you have htop available. So in order to find where in your program you are requesting all that memory, you can open a SWAN terminal with htop running and you can run your notebook cell by cell to check on htop where you see the increment for your notebook process.
I have checked htop in different lines of my code, here are results for “VIRT” :
Closed notebook 0.5GB
Opened nb 1.1GB
Pd DataFrame read from pickle 3.5GB
Train-test split made 7.2GB
Deleted pd DataFrame 4.7
And then, When the train has started VIRT memory goes over 16GB and kernel is killed.
Hi Toni,
do you have the same package versions on your laptop and in SWAN?
This could help in understanding the source of the difference between the two environments.
Cheers, Michał.
Hi Michael, on which packages do you mean?
E.g. sklearn as this seems to be a source of excessive use of the memory.
I have just updated sklearn, pandas and uproot but the problem persists.
This is screenshot just before I delete DF and before I run fit.
Do you think it might be worth asking sklearn support about the memory usage of the logistic regression fit?