Is there a workaround to use languages such as Julia or R in SWAN? There
is package Julia and IJulia maintained by Pere Mato but none of the Software Stacks seem to actually have them?
Dear Jerry,
R and Octave are there. You can create notebooks for them. Concerning Julia I was trying some time ago but there were some difficulties with with the old versions of Julia and the jupyter kernel. Perhaps the situation has changed recently. It can be added if and only if there is use for it.
Cheers,
PereI’m happy to contribute to LCG stack and make Julia part of some release, can you point to me where (CERN gitlab) and how should I test them.
Thanks for you interest. The tool we use to build the stack is https://gitlab.cern.ch/sft/lcgcmake .There is a README with some basic instructions.
Pere
@mato it seems the documentation is a little out-dated (example in REAdME, lcgcmake show configuration should be lcgcmake show config), it would be very helpful if you can clarify something here (and later in the documentation)
- Which branch should I use?
- The quick start instruction does not seem to make lcgcmake environment to work:
$ cd ~/Documents/gitlab/
$ mkdir -p ./lcgcmake_build/opt
$ ARCH=$(uname -m)
$ export PATH=/cvmfs/sft.cern.ch/lcg/contrib/CMake/3.8.1/Linux-${ARCH}/bin:${PATH}
$ git clone https://gitlab.cern.ch/sft/lcgcmake.git
$ export PATH=$PWD/lcgcmake/bin:$PATH
$ lcgcmake configure --compiler=gcc9 --version=dev3python3 --prefix=$HOME/Documents/gitlab/lcgcmake_build/opt
$ lcgcmake install vincia #just as an example
$ lcgcmake run vincia
bash: vincia: command not found
- The way toolchains are configured have changed since this commit, currently, it seems they all use
cmake/toolchain/heptools-common.cmakeand on top of it we do something different, should I make a new file? or add a few lines to existing files like:
LCG_external_package(julia 1.0.4 )
LCG_external_package(Ijulia 1.18.1) #assuming later I can somehow add these to the tarball source
Thanks
- Which branch should I use?
master is good
- The quick start instruction does not seem to make lcgcmake environment to work:
$ cd ~/Documents/gitlab/ $ mkdir -p ./lcgcmake_build/opt $ ARCH=$(uname -m) $ export PATH=/cvmfs/sft.cern.ch/lcg/contrib/CMake/3.8.1/Linux-${ARCH}/bin:${PATH} $ git clone https://gitlab.cern.ch/sft/lcgcmake.git $ export PATH=$PWD/lcgcmake/bin:$PATH $ lcgcmake configure --compiler=gcc9 --version=dev3python3 --prefix=$HOME/Documents/gitlab/lcgcmake_build/opt
Several comments:
- We have not made any release with gcc9, it is still experimental. This is why is trying to build from sources. I would use gcc8 instead of gcc9.
- In principle you do not need to setup a CMake. If the system version is not good enough the lcgcmake command will download a good one.
- Following works nicely for me:
git clone https://gitlab.cern.ch/sft/lcgcmake.git
export PATH=$PWD/lcgcmake/bin:$PATH
mkdir tmp; cd tmp
lcgcmake configure --compiler=gcc8 --version=dev3python3 --prefix=../install
lcgcmake install vincia
$ lcgcmake run vincia bash: vincia: command not found
This is normal because there is no command or executable called vincia. Try for instance
lcgcmake install graphviz
lcgcmake run 'dot -V'
- The way toolchains are configured have changed since this commit, currently, it seems they all use
cmake/toolchain/heptools-common.cmakeand on top of it we do something different, should I make a new file? or add a few lines to existing files like:LCG_external_package(julia 1.0.4 ) LCG_external_package(Ijulia 1.18.1) #assuming later I can somehow add these to the tarball source
Yes, the toolchains have changed. We now include in all the toolchains a common part with all the versions. In particular the one that tried to use heptools-dev3python3.cmake includes heptools-dev-base.cmake. So, the changes with the J=julia/Ijulia versions should go there.
it looks like if I install and then rm -rf tmp/ install/, then try to run lcgcmake configure --compiler=gcc8 --version=dev3python3 --prefix=../install again, it wil error out:
Installing CMake version 3.11.1
CMake Error at .../Documents/gitlab/install/cmake-3.11.1-Linux-x86_64/share/cmake-3.11/Modules/CMakeDetermineCCompiler.cmake:48 (message):
Could not find compiler set in environment variable CC:
.../Documents/gitlab/install/gcc/8.2.0-3fa06/x86_64-centos7/bin/gcc.
Call Stack (most recent call first):
CMakeLists.txt:2 (project)
Something happened to my local environment when the first install configured gcc8?
EDIT: I see it probably has something to do with https://gitlab.cern.ch/jiling/lcgcmake/blob/master/bin/lcgcmake#L370 things I have gcc8
Please do lcgcmake reset if you have deleted by hand some directories.
So julia ends up needing CMake >= 3.4.3 so exporting a 3.8.1 solves the problem and it now builds,
the problem is julia wants a patched LLVM and at the moment remove rpath seems to remove all the runtime libraries:
-- Removed runtime path from ".../install/dev3python3/julia/1.0.4/x86_64-centos7-gcc8-opt/lib/julia/libLLVM-6.0.0.so"
-- Removed runtime path from ".../install/dev3python3/julia/1.0.4/x86_64-centos7-gcc8-opt/lib/julia/libamd.so"
-- Removed runtime path from ".../install/dev3python3/julia/1.0.4/x86_64-centos7-gcc8-opt/lib/julia/libcamd.so"
...
...
should this be fixed by telling lcgcmake to not remove this runtime path or by telling julia to put the library somewhere else?
EDIT: noticed they are actually just moved(soft linked) one directory deep into ...install/dev3python3/x86_64-centos7-gcc8-opt/lib/julia
We do remove in general the rpath to make the build artefacts relocatable. In this case since the libraries are put one level down, we may need to add in the LD_LIBRARY_PATH that is created while sourcing the view setup.sh script the lib/julia directory.
Have a look at cmake/scripts/create_lcg_view_setup_[c]sh.in. We could add section like the one for cuda or tensorflow.
Submitted a PR, this will make julia usable.
I noticed that some packages such as IJulia needs to be installed at compile time(?) as well (due to integration to Jupyter), how should I do this? And how should I install other packages? maybe this but juliaexternal?
I noticed it is possible to install packages on /eos/<home> and tell julia executable to look for packages there, this may be easier
P.S compilation takes forever, is this normal?
We can re-direct Julia’s project path by setting JULIA_DEPOT_PATH, but one issue is that this directory needs to be writable due to recompile cache file and stuff, not sure if there’s a way to separate them.
As for IJulia i.e Jupyter integration, it looks like we need to run a script using Julia:
ENV["XDG_DATA_HOME"]=".../install/dev3python3/x86_64-centos7-gcc8-opt/share/"
import Pkg
Pkg.add("IJulia")
Pkg.build("IJulia")
so that Jupyter kernel will have a ...share/kernels/julia-1.0/kernel.json with:
{
"display_name": "Julia 1.0.4",
"argv": [
"/afs/cern.ch/user/j/jiling/Documents/gitlab/install/dev3python3/julia/1.0.4/x86_64-centos7-gcc8-opt/bin/julia",
"-i",
"--startup-file=yes",
"--color=yes",
"--project=@.",
"/afs/cern.ch/user/j/jiling/.julia/packages/IJulia/gI2uA/src/kernel.jl",
"{connection_file}"
],
"language": "julia",
"env": {},
"interrupt_mode": "signal"
}
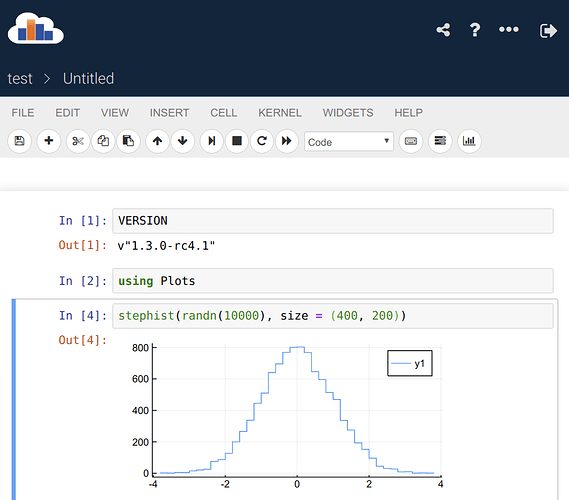
Here’s a way (with some workarounds) to make the official Julia binaries run on SWAN. Open a terminal in SWAN (using the “>_” button) and run
mkdir -p "$HOME/.julia"
mkdir -p "/tmp/`whoami`/.julia/registries"
test -e "$HOME/.julia/registries" || ln -s "/tmp/`whoami`/.julia/registries" "$HOME/.julia/registries"
if [ -z "`command -v julia`" ] ; then
if [ ! -e "$HOME/sw/julia" ] ; then
echo "Installing Julia to \"$HOME/sw/julia\"" >&2
mkdir -p "$HOME/sw/julia" && \
curl "https://julialang-s3.julialang.org/bin/linux/x64/1.3/julia-1.3.0-rc4-linux-x86_64.tar.gz" | \
tar xz --strip-components=1 -C "$HOME/sw/julia"
fi
echo "Adding \"$HOME/sw/julia/bin\" to PATH" >&2
export PATH="$HOME/sw/julia/bin:$PATH"
fi
if [ ! -d "$HOME/.julia/packages/IJulia" ] ; then
echo "Installing IJulia" >&2
export JUPYTER_DATA_DIR="/scratch/`whoami`/.local/share/jupyter"
julia -e 'using Pkg; pkg"add IJulia"'
fi
# Directory "/scratch" does not persist between sessions, reinstall
# Julia Jupyter kernel if necessary:
export JUPYTER_DATA_DIR="/scratch/`whoami`/.local/share/jupyter"
julia -e 'using IJulia; installkernel("julia", env=merge(Dict("JULIA_NUM_THREADS"=>"4"), Dict(ENV)))'
Then reload the SWAN main web page, and:
There’s two tricks here:
-
EOS really doesn’t seem to be the Julia package registry - I guess because EOS isn’t fully Posix-compatible in all respects. So
~/.julia/registriesneeds to be redirected to/tmp/.... Not a great loss - there’s many small files in there, so it’s faster on/tmpanyhow, and the registry is only needed for package installation and updates. And it’s quick to regenerate. -
SWAN allows custom Jupyter kernels to be installed in
/scratch/USERNAME/.local/share/jupyter- but since the contents of/scratchdon’t survive the session, the kernel needs to be reinstalled in a new session (by setting$JUPYTER_DATA_DIRand runningusing IJulia; installkernel("julia"))
It would be really great though if CERN IT could be persuaded to make the latest stable Julia version part of the software distribution (compiled for CentOS-7, so that Cxx.jl works properly, etc.).
I’m sure with official backing, one could also implement a solution for the problems with the registry. And the Jupyter kernel could come pre-installed (along with IJulia itself).
To use UpROOT.jl on SWAN, you first need to install the Python uproot package. You can install it into “$HOME/.local/lib/python3.6/site-packages” via
pip install --user uproot
Python should find it there automatically. Also, it’s necessary to carry a whole bunch of environment variables that are set on the SWAN shell over into the notebooks, by setting your Julia Jupyter kernel up like this:
julia -e 'using IJulia; installkernel("julia", env=merge(Dict("JULIA_NUM_THREADS"=>"4"), Dict(ENV)))'
Setting the number of Julia threads is optional, of course, but you need to preserve ENV, there are a lot of custom python, library and others paths set up on the SWAN shell. Take a look at
cat "/scratch/`whoami`/.local/share/jupyter/kernels/python3/kernel.json"
to see how it’s done for the Python Jupyter kernel (pretty monstrous).
Important: Run a SWAN instance with Python3 if you plan to install Julia packages like UpROOT, PyPlot, PyCall, etc. - if the system Python is Python2, PyCall/Conda will install a whole new Conda envinronment into “.julia/conda” and EOS won’t be happy about those many many small files.
I have upgraded the version of julia to 1.6 in the LCG stack (Bleeding Edge for the time being). From your workarounds the minimal set of commands to execute at the terminal are:
export JUPYTER_DATA_DIR=$HOME/.local/share/jupyter
mkdir -p /scratch/`whoami`/.julia/registries
ln -s "/scratch/`whoami`/.julia/registries" "$HOME/.julia/registries"
julia -e 'import Pkg; Pkg.add("IJulia")'
julia -e 'using IJulia; installkernel("julia", env=merge(Dict("JULIA_NUM_THREADS"=>"4"), Dict(ENV)))'
Then I can start a julia notebook and execute it. If I re-start the session I do not need to execute any command at the termninal. I guess until a new version is deployed.
Not really that I can re-start the session. I need at least to re-create registries directory with
mkdir -p /scratch/`whoami`/.julia/registries
for the moment being it hangs after Installing known registries into `~/.julia for me
edit: nvm, seems to be just that the storage is outrageously slow…
I tried to halt notebook and logout then login, opening notebook worked just fine. looks like $HOME is just the user /eos root directory and if that’s where .julia/ lives, everything works perfectly fine.
Yes, this is because CERNBox (EOS) is extremely slow when you have create many small files. This is the reason to move the registries to /scratch/`whoami`/.julia/registries and create a link in ~/.julia/registries.
Hi Mato,
Is it possible to host a read-only /registries somewhere for all the user to use and just git pull every 12 hours or so? If this is doable, here’s what we need and how user’s Julia would interact:
- the general registry is just a git repo, say we have it cloned as
$JD_PATH/registries/General - the only thing user then needs to do is to
export JULIA_DEPOT_PATH=":$JD_PATH", the empty before:allows the default paths to populate first.
here’s more detailed explanation FYI:
2. any Julia session would populate theDEPOT_PATH(just type this after opening up the REPL) according to ENV variableJULIA_DEPOT_PATH, when this ENV variable is empty, it puts~/.julia/and other two default pathes in theDEPOT_PATH.
3. user would only ever write to the first path (DEPOT_PATH[1]), this is where package source files exist and precompile binary files live, but anything after are read-only.