In Using a conda environment I mentioned that I’d previously been able to use the “Environment script” section of the SWAN startup to configure a custom Jupyter kernel. In my case, I simply installed a conda environment into my $SCRATCH_HOME directory at startup to include a newer version of Python and a few key packages. @dalvesde said that he would be interested in the approach, since there is some planning work around “environment scripts” and even providing conda environments, and that it would be good to get an example of what you can already do if you know your way around the ecosystem.
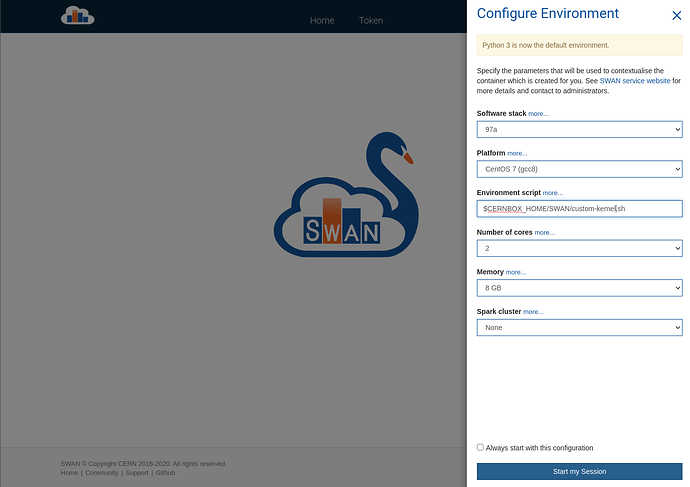
In that light, let’s go step-by-step the approach I took. First, I created a script in my homespace that I can reference in the SWAN configurator screen (the one before your Jupyter environment has been loaded). For the record, as somebody who doesn’t know the CERN infrastructure too well yet, I originally was creating these files by starting SWAN and modifying them with nano in the web terminal. I realise now that I could have just logged on to lxplus.cern.ch and gone to /eos/home-p/pelson.
The steps that I needed:
- Drop all of the special environment variables. Since the SWAN environment either sets or inherits (likely the latter) a bunch of environment variables which would pollute a clean environment. These include the PYTHONHOME, PYTHONPATH and LD_LIBRARY_PATH environment variables.
unset PYTHONHOME
unset PYTHONPATH
unset LD_LIBRARY_PATH
unalias python &>/dev/null
- Install conda. Using the age-old trick of downloading miniconda, you can quickly get yourself setup with a fresh conda installation. In 2020 though there is a much quicker way of bootstrapping a conda environment for linux called micromamba (the steps are pretty much the same if you want to use conda):
export MAMBA_ROOT_PREFIX=${SCRATCH_HOME}/mamba
MICROMAMBA=${MAMBA_ROOT_PREFIX}/bin/micromamba
if [ ! -f "${MICROMAMBA}" ]
then
mkdir -p $(dirname ${MICROMAMBA})"
cd ${MAMBA_ROOT_PREFIX}
wget -qO- https://micromamba.snakepit.net/api/micromamba/linux-64/latest | tar -xvj bin/micromamba
fi
- Create an environment.
KERNEL_NAME=my-custom-kernel # Could equally be python3 if you want to replace the default kernel
ENVIRONMENT_PREFIX=$MAMBA_ROOT_PREFIX/envs/$KERNEL_NAME
$MICROMAMBA create -p $ENVIRONMENT_PREFIX ipykernel the-packages-you-want
- Register the environment as a jupyter kernel. This step allows SWAN’s Jupyter interface to find the environment you’ve just installed. If you aren’t using a Python kernel (e.g. you want a clang kernel, or an R/Julia/etc. kernel) then install the necessary kernel package with conda/mamba above and follow the steps to register that kernel. For Python it is:
KERNEL_PREFIX=$SCRATCH/.local # Actually you can derive this from the KERNEL_DIR environment variable.
$ENVIRONMENT_PREFIX/bin/python -m ipykernel install --prefix=${KERNEL_PREFIX} --name ${KERNEL_NAME}
- Workaround the 60s limit of the environment script. The SWAN environment script has a timeout of 60 seconds (sounds like this is going to be extended). Conda takes a little longer than this to create an environment (mamba’s performance is much better, and mostly falls below the 60s limit if your environment isn’t huge). To do this, we simply run the conda steps (2 & 3) in a background process, and in the meantime “trick” jupyter into thinking it has a kernel definition which it can use immediately. This means that the web UI shows the kernel in the drop-down when creating a new notebook or modifying the kernel of an existing one, but that by the time you do this, you will want your background script to have completed, otherwise you will get some ugly errors when trying to start the kernel.
# Create a quick kernel definition, even though the installation hasn't yet finished.
# This will allow the SWAN interface to at least recognise its existence whilst the
# installation is taking place.
mkdir -p $KERNEL_PREFIX/share/jupyter/kernels/${KERNEL_NAME}
cat <<EOF > $KERNEL_PREFIX/share/jupyter/kernels/${KERNEL_NAME}/kernel.json
{
"argv": [
"${ENVIRONMENT_PREFIX}/bin/python",
"-m",
"ipykernel_launcher",
"-f",
"{connection_file}"
],
"display_name": "${KERNEL_NAME}",
"language": "python"
}
EOF
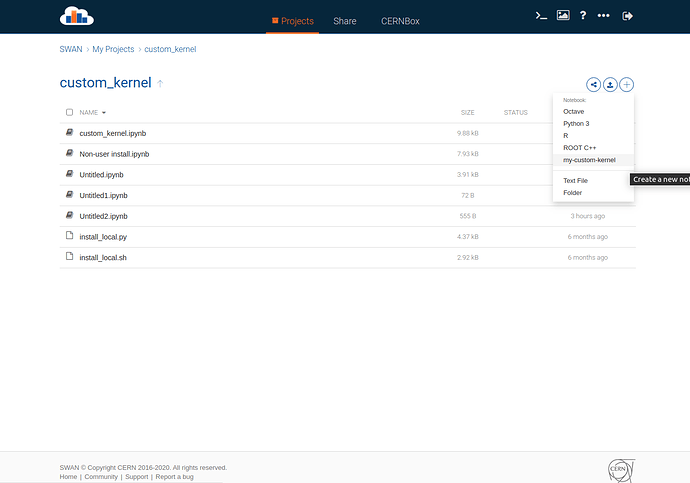
A few screenshots to show the workfolow:
Caveats
To be completely clear, I want to stress that I consider this a bit of a hack and I do not endorse doing this. I’m posting this in the interest of transparency to demonstrate what is possible today in the hope that it will be useful to the SWAN maintainers in the hope that they can integrate the custom workflow into the SWAN service. One example of the costs of the approach is the amount of data which is being downloaded/installed per SWAN session (the order of 500MB for a basic Python environment with conda) - this is the kind of thing that we could have a common cache for such that all users on SWAN can re-use the same downloaded binary artefact.
@dalvesde I’d be very happy to be involved in any requirements gathering / prototype evaluation when you start planning/implementing custom environments in SWAN. If you have any questions, or just want to chat about these things, just drop me a line  .
.